Overview
tajima can be used to calculate Tajima's D across a sliding window or using bins. For an explanation of what Tajima's D is, see this excellent video by Mohamad Noor.
In order for a SNP to be incorporated in the calculation, it must:
- Have an allele frequency greater than 0 and less than 1.
- Be biallelic.
- Be a SNP.
- Be a diploid site.
Usage
Parameters:
- window-size - Size of window from in which to calculate Tajima's D.
- step-size - Size of step taken.
- --sliding - Fluidly slide along genome, capturing every window of a given
window-size. Equivelent tostep-size= 1; - --no-header - Outputs results without a header.
- --extra - Adds on
filename,window-sizeandstep-sizeas additional columns. Useful for comparing different files / parameters.
Tip
You can specify window-size and step-size using commas or scientific notation (e.g. 1,000,000 or 1E7).
Output
tajima will output the following columns:
- CHROM
- BIN_START - Starting interval position inclusive.
- BIN_END - Ending interval position not inclusive.
- N_Sites - Number of sites used to calculate Tajima's D.
- N_SNPs - Number of SNPs present in interval. Certain sites are excluded.
- TajimaD - Tajima's D calculation.
If you optionally specify --extra, the following columns will also be included in output:
- filename
- window_size
- step_size
Examples
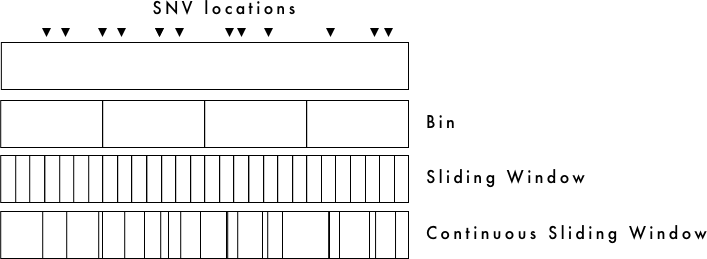
The figure above illustrates the types of windows over which Tajima's D can be calculated.
Bin calculation
If you set the window-size and step-size as the same value, the bins will not overlap. This is depicted in the figure above as 'Bin'.
vk tajima 1,000,000 1,000,000 <vcf>
The code above will calculate Tajima's D using 100,000 bp bins across the genome.
| CHROM | BIN_START | BIN_END | N_Sites | N_SNPs | TajimaD |
|---|---|---|---|---|---|
| I | 0 | 1000000 | 24 | 8 | -0.344142 |
| I | 1000000 | 2000000 | 47 | 20 | 0.666153 |
| I | 2000000 | 3000000 | 34 | 18 | 0.418091 |
| I | 3000000 | 4000000 | 22 | 10 | -0.676877 |
| I | 4000000 | 5000000 | 11 | 4 | -0.652344 |
| I | 5000000 | 6000000 | 8 | 2 | -0.498306 |
| I | 7000000 | 8000000 | 8 | 4 | -0.537028 |
Sliding window
When run, the code below will calculate Tajima's D across a 100,000 bp sliding window that moves 1,000 bp with each iteratino.
vk tajima 100,000 1,000 <vcf>
| CHROM | BIN_START | BIN_END | N_Sites | N_SNPs | TajimaD |
|---|---|---|---|---|---|
| I | 6000 | 106000 | 2 | 1 | -0.740994 |
| I | 7000 | 107000 | 2 | 1 | -0.740994 |
| I | 8000 | 108000 | 2 | 1 | -0.740994 |
| I | 9000 | 109000 | 2 | 1 | -0.740994 |
| I | 10000 | 110000 | 2 | 1 | -0.740994 |
| I | 11000 | 111000 | 2 | 1 | -0.740994 |
Continous sliding window
When run, the code below will calculate Tajima's D across a 100,000 bp sliding window that captures every unique bin of variants that fall within 100,000 bp of one another.
vk tajima 1E5 --sliding <vcf>
| CHROM | BIN_START | BIN_END | N_Sites | N_SNPs | TajimaD |
|---|---|---|---|---|---|
| I | 0 | 100000 | 2 | 1 | -0.740994 |
| I | 90777 | 190777 | 2 | 2 | -0.0333856 |
| I | 154576 | 254576 | 2 | 1 | 0.690099 |
| I | 207871 | 307871 | 2 | 1 | -0.740994 |
| I | 263709 | 363709 | 2 | 1 | -0.740994 |
| I | 321321 | 421321 | 2 | 1 | -0.740994 |
| I | 294407 | 394407 | 3 | 1 | -0.740994 |
| I | 391250 | 491250 | 3 | 2 | -0.110617 |